Author
Author  Correspondence author
Correspondence author
Maize Genomics and Genetics, 2024, Vol. 15, No. 4 doi: 10.5376/mgg.2024.15.0017
Received: 30 May, 2024 Accepted: 02 Jul., 2024 Published: 18 Jul., 2024
Zhu S.Z., and Wang W., 2024, Teosinte and its role in maize genetic enhancement, Maize Genomics and Genetics, 15(4): 171-181 (doi: 10.5376/mgg.2024.15.0017)
This study explores the crucial role of teosinte in the genetic enhancement of maize. As the wild ancestor of modern maize, teosinte possesses rich genetic diversity and novel alleles that were lost during domestication, making it an important genetic resource for maize improvement. Research indicates that teosinte alleles can enhance various agronomic traits in maize, such as yield, stress resistance, and nutritional quality. For example, the introduction of the UPA2 allele from teosinte has significantly increased maize yield under high-density planting conditions by altering plant architecture. Additionally, teosinte's genetic diversity includes strong alleles that control kernel composition traits, such as starch, protein, and oil content, which can improve the nutritional value of maize. The integration of archaeological and molecular evidence has significantly advanced the understanding of the teosinte-maize relationship, highlighting the potential of teosinte in modern maize breeding programs. Techniques such as hybridization and backcrossing, marker-assisted selection (MAS), genomic selection (GS), and CRISPR/Cas9 gene editing allow researchers to effectively utilize teosinte's genetic diversity to develop superior maize varieties with improved agronomic traits and resilience to environmental stresses. Despite the genetic barriers, breeding difficulties, and regulatory and ethical issues associated with using teosinte for maize improvement, these challenges can be overcome through global collaboration and germplasm conservation. In the future, advanced genomic tools and techniques, the exploration of new potential traits from teosinte, and the integration of teosinte into sustainable agriculture practices will fully realize its potential in maize genetic enhancement, leading to the development of superior maize varieties that meet the demands of modern agriculture and contribute to global food security.
1 Introduction
Maize (Zea mays ssp. mays) is one of the most important cereal crops globally, and its domestication from its wild ancestor, teosinte (Zea mays ssp. parviglumis), is a remarkable example of plant evolution driven by human selection. The domestication process, which began approximately 9 000 years ago, involved significant morphological and genetic changes, transforming teosinte's small, hard kernels into the large, soft kernels of modern maize (Sahoo et al., 2019). This transformation was facilitated by the selection of favorable traits, such as reduced branching and increased kernel size, which were controlled by a relatively small number of major loci (Liu et al., 2019). However, this process also led to a reduction in genetic diversity due to domestication and selection bottlenecks (Warburton et al., 2011).
Teosinte, the wild progenitor of maize, remains a valuable genetic resource for maize improvement due to its greater genetic diversity and the presence of novel alleles that were lost during domestication (Karn et al., 2011; Fang et al., 2019). Teosinte harbors alleles that can enhance agronomic traits such as yield, stress resistance, and nutritional quality (Sahoo et al., 2021). For instance, the introgression of teosinte alleles has been shown to improve high-density maize yields by altering plant architecture (Tian et al., 2019). Additionally, teosinte's genetic diversity includes alleles that contribute to kernel composition traits, such as increased oil and protein content, which are beneficial for both human and animal nutrition (Karn et al., 2011). The genetic basis of these traits has been elucidated through various studies, highlighting the potential of teosinte in modern maize breeding programs (Hubbard et al., 2002; Liu et al., 2019).
This study aims to provide a comprehensive overview of the role of teosinte in maize genetic enhancement. It will explore the genetic and phenotypic differences between maize and teosinte, the potential of teosinte alleles to improve modern maize, and the mechanisms by which these alleles can be integrated into maize breeding programs. By examining the current status and future prospects of utilizing teosinte in maize improvement, this study seeks to underscore the importance of conserving and studying wild germplasm to ensure the continued evolution and resilience of maize in the face of changing environmental conditions and agricultural demands.
2 Teosinte: Biological and Genetic Background
2.1 Taxonomy and species of teosinte
Teosinte, the wild ancestor of modern maize (Zea mays L.), belongs to the genus Zea and comprises several species and subspecies. The primary taxa include Zea mays ssp. parviglumis, Zea mays ssp. mexicana, Zea diploperennis, Zea luxurians, Zea perennis, Zea mays ssp. huehuetenangensis, Zea vespertilio, and Zea nicaraguensis. These species are distributed from northern Mexico to Costa Rica, with the Mexican annuals Zea mays ssp. parviglumis and Zea mays ssp. mexicana showing a wide distribution in Mexico, while other species have more restricted ranges (González et al., 2018; Rivera-Rodríguez et al., 2023).
2.2 Morphological characteristics
Teosinte exhibits distinct morphological traits compared to domesticated maize. It typically has a more branched structure with multiple stalks and smaller ears covered by a few layers of husks. The leaves of teosinte are generally narrower and longer than those of maize. The husk traits, including husk length, width, and the number of husk layers, show significant variation between teosinte and maize, with teosinte having fewer and smaller husk layers (Fu et al., 2019; Zhang et al., 2021). Additionally, teosinte kernels are encased in a hard fruitcase, unlike the exposed kernels of maize (Chen et al., 2020).
2.3 Genetic diversity in teosinte
Teosinte harbors a high level of genetic diversity, which is crucial for its adaptation to various environmental conditions. Studies have shown that Zea mays ssp. parviglumis and Zea mays ssp. mexicana exhibit the highest levels of genomic diversity among the teosinte taxa (Rivera-Rodríguez et al., 2023). This genetic diversity is influenced by local adaptation to environmental factors such as temperature and soil phosphorus concentration, as well as historical climate fluctuations during the Holocene and Pleistocene (Aguirre-Liguori et al., 2019; Gasca-Pineda et al., 2023). The genetic structure of teosinte populations reveals significant differentiation between and within subspecies, driven by both contemporary and historical environmental factors (Gasca-Pineda et al., 2023). This diversity makes teosinte a valuable genetic resource for maize breeding programs aimed at improving traits such as stress tolerance and nutritional content (Figure 1) (Zavala-López et al., 2018; Xu et al., 2019).
|
Figure 1 Metabolome divergence between maize and teosinte (Adopted from Xu et al., 2019) Image caption: (A): Classification of metabolites that have annotated structures; (B): Principal component analysis (PCA) of the maize and teosinte accessions with all metabolites (Adopted from Xu et al., 2019) |
3 Historical Perspectives
3.1 Early studies on teosinte-maize relationships
The relationship between teosinte and maize has been a subject of scientific inquiry for many years. Early studies focused on the morphological differences and similarities between the two plants. Teosinte, the wild ancestor of maize, exhibits a significantly different phenotype compared to modern maize, despite their genetic similarities (Zobrist et al., 2021). Initial genetic studies revealed that a small number of major loci could explain a large portion of the phenotypic changes observed during maize domestication (Liu et al., 2019). These findings supported the hypothesis that selective breeding and domestication led to the remarkable transformation of teosinte into modern maize.
3.2 Archeological evidence
Archeological evidence has played a crucial role in understanding the domestication process of maize from teosinte. Fossil records and ancient maize cobs found in archeological sites provide insights into the early stages of maize domestication. These records indicate that teosinte was first domesticated around 10 000 years ago in the Balsas River Valley of southern Mexico (Adhikari et al., 2021). The archeological findings suggest that early agricultural communities selectively bred teosinte for desirable traits, leading to the gradual transformation into maize. This evidence underscores the importance of teosinte as a genetic reservoir that contributed to the development of modern maize varieties.
3.3 Molecular evidence
Molecular studies have further elucidated the genetic relationship between teosinte and maize. Advances in genomic technologies have allowed researchers to identify specific genes and quantitative trait loci (QTLs) that differentiate teosinte from maize. For instance, single-molecule long-read sequencing has revealed extensive genomic and transcriptomic variation between maize and its wild relative teosinte (Li et al., 2021). This study identified 70 044 nonredundant transcript isoforms and constructed a draft genome of teosinte, providing a valuable resource for maize breeding programs.
Additionally, molecular evidence has shown that teosinte harbors unique alleles that can enhance modern maize varieties. For example, the UPA2 allele, which reduces leaf angle and improves high-density maize yields, originated from teosinte but was lost during domestication. Introgressing such wild alleles into modern maize hybrids can enhance agronomic traits and yield potentia (Tian et al., 2019).
Overall, the integration of archeological and molecular evidence has significantly advanced our understanding of the teosinte-maize relationship, highlighting the potential of teosinte as a valuable genetic resource for maize improvement.
4 Genetic Contributions of Teosinte to Maize
4.1 Genetic loci associated with domestication
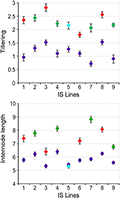
Teosinte, the wild ancestor of maize, has played a crucial role in the domestication and genetic enhancement of maize. The genetic architecture of teosinte and maize has been extensively studied to understand the loci associated with domestication. One significant locus is the teosinte branched1 (tb1) gene, which has been shown to have large effects on plant architecture and ear morphology. The tb1 gene is involved in the plant's response to environmental conditions, influencing the development of long or short branches, which was a key factor in maize domestication (Figure 2) (Studer et al., 2012). Additionally, other loci such as UPA1 and UPA2, which regulate plant architecture, have been identified. These loci contribute to the upright plant architecture that facilitates dense planting, a trait beneficial for modern agriculture (Tian et al., 2019).
|
Figure 2 Phenotypic Means of Introgressed Segments from Various Zea Species (Adopted from Studer et al., 2012) Image caption: Points are shaded on the basis of taxonomic origin of the tb1 introgressed segment: (purple) Zea mays ssp. mays control populations, (blue) Z. diploperennis, (red) Z. mays ssp. parviglumis, and (green) Z. mays ssp. Mexicana; Error bars represent the standard error for each genotypic class. The x-axis shows the introgression segments; the y-axis shows trait means (Adopted from Studer et al., 2012) |
4.2 Traits inherited from teosinte
Several important traits have been inherited from teosinte, contributing to the genetic diversity and adaptability of modern maize. For instance, teosinte harbors stronger alleles for kernel composition traits, including starch, protein, and oil content, which can be exploited for the improvement of these traits in maize (Karn et al., 2017). Another inherited trait is the narrow plant architecture conferred by the UPA2 allele, which enhances high-density maize yield (Figure 3) (Tian et al., 2019). Furthermore, teosinte has contributed to the morphological diversification of maize, with traits such as ear structure and shattering being influenced by genes like ramosa1 (ra1) and zagl1 (Weber et al., 2008). These traits have been crucial in the adaptation and improvement of maize for various agricultural practices..
|
Figure 3 Positional cloning of UPA2A (Adopted from Tian et al., 2019) Image caption: (A): QTL mapping for middle leaf angle in the maize–teosinte BC2S3 population; LOD, logarithm of odds. UPA2 and UPA1 are the largest- and second-largest leaf angle QTL, respectively. The dashed gray line at LOD of 5 indicates the threshold of claiming significant QTLs. (B) Gross morphologies of UPA2-NILW22 and UPA2-NIL8759. The white arrows indicate the lower, middle, and upper leaves in which leaf angle was scored. Scale bars, 20 cm. (C) Comparison of leaf angle in lower, middle, and upper leaves between UPA2-NILW22 and UPA2-NIL8759. (D) Scanning electron microscopy analysis of the ligular region of UPA2-NILW22 and UPA2-NIL8759. The ligular band and the mature auricle region are indicated by white dashed lines. Scale bars, 3 mm (top) and 500 mm (bottom). (E) Comparison of the width of the ligular band and auricle margin between UPA2-NILW22 and UPA2-NIL8759. (F) Cross-sections of the mature ligular region of UPA2-NILW22 and UPA2-NIL8759. Top shows the abaxial side and bottom shows the adaxial side. Scale bars, 100 mm; (G) Comparison of number of the abaxial sclerenchyma cell layers (left) and the adaxial sclerenchyma cell layers (right) between UPA2-NILW22 and UPA2-NIL8759. (H) Location of UPA2 on maize chromosome 2; CEN, centromere; (I) Fine mapping of UPA2 using an NIL population (n = 3180). The number of recombinants between adjacent markers is indicated below the linkage map. (J) Progeny testing of recombinants delimited UPA2 to a 240-bp noncoding region (red lines). The graphical genotypes of the five critical recombinants are shown on the left. White, black, and gray segments indicate regions homozygous for W22, regions homozygous for 8759, and heterozygous regions, respectively. The bar graphs on the right compare middle leaf angle between homozygous recombinants and homozygous nonrecombinants within each recombinant-derived F3 family. Black and white bars represent homozygous progenies that inherited the 8759 and W22 chromosome from the parental recombinant, respectively. The 240-bp region of UPA2 is located 9540 bp upstream of the start codon (ATG) of GRMZM2G102059 (ZmRAVL1). Pink and gray regions indicate the exon and untranslated regions (UTR), respectively; Values are means±SD. **P<0.01 (Student’s t test); N.S., not significant (Adopted from Tian et al., 2019) |
4.3 Modern genetic analysis of teosinte
Modern genetic analysis techniques have provided deeper insights into the genetic contributions of teosinte to maize. Single-molecule long-read sequencing has revealed extensive genomic and transcriptomic variation between maize and teosinte, identifying numerous nonredundant transcript isoforms and providing a robust gene classifier for complex genomes (Li et al., 2021). Additionally, association mapping studies have identified significant associations between specific genes and trait variations in teosinte, suggesting new putative causative relationships for domestication traits (Weber et al., 2008). These advanced genetic analyses have furthered our understanding of maize domestication and provided valuable resources for utilizing teosinte germplasm in maize breeding (Huang et al., 2016).
In summary, the genetic contributions of teosinte to maize are profound, with key loci such as tb1 and UPA2 playing significant roles in domestication. Traits inherited from teosinte, including kernel composition and plant architecture, have been crucial for maize improvement. Modern genetic analysis techniques continue to uncover the complex genetic architecture of teosinte, providing valuable insights and resources for future maize breeding efforts.
5 Techniques for Utilizing Teosinte in Maize Breeding
5.1 Hybridization and backcrossing
Hybridization and backcrossing are traditional techniques used to introduce desirable traits from teosinte into maize. By crossing teosinte with elite maize lines, researchers can create hybrids that combine the beneficial traits of both species. Subsequent backcrossing with maize helps to retain the desired traits while maintaining the overall genetic makeup of maize. For instance, teosinte has been used to develop backcross inbred lines (BILs) that exhibit resistance to pests like the red flour beetle and improved plant architecture traits (Joshi et al., 2021; Adhikari et al., 2022). These BILs are evaluated for various agronomic traits, and quantitative trait loci (QTLs) are mapped to identify genomic regions associated with these traits (Joshi et al., 2021; Adhikari et al., 2022).
5.2 Marker-assisted selection (MAS)
Marker-Assisted Selection (MAS) is a technique that uses molecular markers to select plants with desirable traits during the breeding process. This method accelerates the breeding cycle by allowing for the early identification of plants carrying the desired genes. In the context of teosinte and maize, MAS has been employed to introgress traits such as pest resistance and improved plant architecture. For example, SSR markers have been used to map QTLs governing traits like plant height, leaf length, and ear number in teosinte-introgressed maize populations (Kumar et al., 2020; Adhikari et al., 2022). These markers facilitate the selection of superior lines that can be used in further breeding programs.
5.3 Genomic selection (GS)
Genomic Selection (GS) is an advanced breeding technique that uses genome-wide markers to predict the performance of breeding lines. This method allows for the selection of plants with the best genetic potential for multiple traits simultaneously. In maize breeding, GS can be particularly useful for incorporating complex traits from teosinte, such as stress resistance and yield improvement. Studies have shown that introgressed teosinte alleles can enhance maize's adaptability to different environmental conditions, making GS a valuable tool for developing resilient maize varieties (Calfee et al., 2021; Adhikari et al., 2022).
5.4 CRISPR/Cas9 and gene editing
CRISPR/Cas9 and other gene-editing technologies offer precise methods for introducing or modifying specific genes in the maize genome. These techniques can be used to directly edit genes identified in teosinte that confer beneficial traits. For example, CRISPR/Cas9 has been used to create waxy corn hybrids with higher yields compared to those produced by traditional breeding methods (Gao et al., 2020). Additionally, gene editing can be employed to introgress specific alleles from teosinte that have been lost during maize domestication, thereby enhancing traits like plant architecture and yield under high-density planting conditions (Tian et al., 2019).
By leveraging these techniques, researchers can effectively utilize the genetic diversity of teosinte to enhance maize breeding programs, leading to the development of superior maize varieties with improved agronomic traits and resilience to environmental stresses.
6 Case Studies of Teosinte Utilization
6.1 Drought tolerance
Teosinte has been instrumental in enhancing drought tolerance in maize through various genetic approaches. For instance, the development of hybrid models using artificial intelligence techniques has shown promise in predicting drought tolerance indices in teosinte introgressed maize lines. The genetic algorithm-based support vector machine (SVM-GA) model, in particular, has demonstrated high potential in forecasting drought tolerance and stress tolerance indices, making it a valuable tool for improving drought resilience in maize (Kumar et al., 2022). Additionally, the TCP family genes, specifically ZmTCP42, have been identified to play a significant role in drought tolerance. Overexpression of ZmTCP42 in Arabidopsis has validated its function in enhancing drought tolerance, suggesting its potential application in maize breeding programs (Ding et al., 2019).
6.2 Disease resistance
Teosinte-derived alleles have also been utilized to confer multiple disease resistances in maize. A notable example is the teosinte-derived allele of the Mexicana lesion mimic 1 (ZmMM1) gene, which has been shown to provide resistance to northern leaf blight (NLB), gray leaf spot (GLS), and southern corn rust (SCR). The strong multiple disease resistance (MDR) conferred by this allele is linked to polymorphisms in the 3' untranslated region of the ZmMM1 gene, leading to increased accumulation of the ZmMM1 protein. This discovery not only aids in developing broad-spectrum and durable disease resistance but also enhances our understanding of the molecular mechanisms underlying MDR (Wang et al., 2021).
6.3 Yield improvement
Teosinte has contributed to yield improvement in maize, particularly under high-density planting conditions. The identification and introgression of the UPA2 allele from teosinte, which regulates upright plant architecture, have been shown to enhance maize yields in densely planted fields. This allele, which was lost during maize domestication, has been reintroduced to modern hybrids, resulting in improved agronomic characteristics and higher yields under high-density planting conditions (Tian et al., 2019). Additionally, the identification of a new QTL underlying seminal root number in a maize-teosinte population has provided insights into improving root architecture, which is crucial for water and nutrient acquisition, ultimately contributing to yield improvement (Wang et al., 2023).
6.4 Nutritional quality enhancement
Teosinte has also been explored for its potential to enhance the nutritional quality of maize. Although specific studies on nutritional quality enhancement were not directly cited in the provided data, the overall genetic diversity and unique traits of teosinte, such as its ability to form aerenchyma under flooding conditions, suggest that it holds promise for improving various agronomic traits, including nutritional quality. The introgression of beneficial alleles from teosinte into maize can potentially lead to the development of maize varieties with enhanced nutritional profiles, thereby addressing both yield and quality aspects of maize production (Mano et al., 2006; Mano and Omori, 2007; 2013).
In summary, teosinte has played a crucial role in maize genetic enhancement, contributing to improved drought tolerance, disease resistance, yield, and potentially nutritional quality. The utilization of teosinte-derived alleles and genetic resources continues to offer valuable opportunities for advancing maize breeding programs and addressing global agricultural challenges.
7 Challenges and Limitations
The integration of teosinte into maize genetic enhancement presents several challenges and limitations. These can be broadly categorized into genetic barriers, breeding difficulties, and regulatory and ethical issues.
7.1 Genetic barriers
One of the primary challenges in utilizing teosinte for maize improvement is the genetic incompatibility between the two species. The presence of specific loci such as Teosinte crossing barrier1 (Tcb1) restricts hybridization with maize, making it difficult to crossbreed the two species effectively (Evans et al., 2001). Additionally, the extensive genomic and transcriptomic variation between maize and teosinte further complicates the integration of beneficial traits from teosinte into maize (Li et al., 2021). The genetic architecture of teosinte, which includes a high degree of genetic diversity and unique alleles, poses significant barriers to the straightforward transfer of traits (Karn et al., 2017; Yang et al., 2019).
7.2 Breeding difficulties
Breeding teosinte with modern maize varieties is fraught with practical difficulties. The differences in plant and inflorescence architecture between maize and teosinte, controlled by loci such as teosinte branched1 (tb1), result in phenotypic traits that are not always desirable in modern agricultural contexts. Moreover, the genetic background of teosinte can affect the expression of these traits, making it challenging to predict and control the outcomes of breeding programs. The process of backcrossing and selecting for desirable traits is labor-intensive and time-consuming, often requiring multiple generations to achieve stable and beneficial hybrids (Karn et al., 2017).
7.3 Regulatory and Ethical Issues
The use of wild relatives like teosinte in crop improvement also raises several regulatory and ethical concerns. The introduction of wild alleles into commercial maize varieties may be subject to stringent regulatory scrutiny to ensure that the resulting crops are safe for consumption and do not pose environmental risks (Sahoo et al., 2021). Additionally, there are ethical considerations related to the conservation of wild species and the potential impacts of their domestication on biodiversity (Sahoo et al., 2021). The use of genetic modification techniques to incorporate teosinte traits into maize further complicates the regulatory landscape, as these methods are often subject to additional legal and public acceptance hurdles (Tian et al., 2019).
In summary, while teosinte offers valuable genetic resources for maize improvement, the challenges and limitations associated with genetic barriers, breeding difficulties, and regulatory and ethical issues must be carefully navigated to realize its full potential in agricultural applications.
8 Future Directions
8.1 Advanced genomic tools and techniques
The advancement of genomic tools and techniques holds significant promise for the genetic enhancement of maize using teosinte. Single-molecule long-read sequencing has revealed extensive genomic and transcriptomic variations between maize and teosinte, providing a robust resource for maize breeding (Li et al., 2021). The development of state-of-the-art bioinformatics pipelines, such as DenovoAS_Finder, allows for accurate annotation of teosinte transcriptomes without a complete reference genome, facilitating the identification of beneficial alleles (Li et al., 2021). Additionally, the comprehensive transcriptome sequencing of teosinte accessions has identified numerous unigenes with strong selection signals, which can be targeted for maize improvement (Huang et al., 2016). These advanced genomic tools will enable more precise and efficient utilization of teosinte genetic resources in maize breeding programs.
8.2 Potential new traits from teosinte
Teosinte harbors a wealth of genetic diversity that can be harnessed to introduce new traits into maize. For instance, teosinte alleles have been shown to improve kernel composition traits, such as starch, protein, and oil content, which are crucial for enhancing the nutritional value of maize (Karn et al., 2017). Moreover, the identification of the UPA2 allele from teosinte, which confers upright plant architecture, has demonstrated potential for increasing maize yields under high-density planting conditions (Tian et al., 2019). The exploration of teosinte's genetic diversity can lead to the discovery of novel traits that enhance maize's adaptability, resilience, and productivity.
8.3 Integrating teosinte in sustainable agriculture
Integrating teosinte into sustainable agriculture practices can significantly contribute to the resilience and adaptability of maize. Teosinte possesses diverse alleles for resistance to abiotic and biotic stresses, which can be introgressed into maize to enhance its stress tolerance (Sahoo et al., 2021). The development of teosinte-derived maize lines has shown significant diversification in agronomic traits, yield, and adaptation, indicating the potential of teosinte in improving maize's performance under various environmental conditions (Sahoo et al., 2021). By leveraging teosinte's genetic resources, sustainable agriculture practices can be developed to ensure food security and environmental sustainability.
8.4 Global collaborations and germplasm conservation
Global collaborations and germplasm conservation are essential for the effective utilization of teosinte in maize genetic enhancement. The conservation of teosinte germplasm is crucial for maintaining its genetic diversity and ensuring its availability for future breeding programs (Sahoo et al., 2021). Collaborative efforts among researchers, breeders, and conservationists can facilitate the exchange of knowledge, resources, and technologies, promoting the sustainable use of teosinte in maize improvement. Additionally, the establishment of international germplasm repositories and databases can enhance the accessibility and utilization of teosinte genetic resources, fostering global efforts in maize genetic enhancement (Sahoo et al., 2021).
By focusing on these future directions, the potential of teosinte in maize genetic enhancement can be fully realized, leading to the development of superior maize varieties that meet the demands of modern agriculture and contribute to global food security.
9 Concluding Remarks
Teosinte, the wild ancestor of modern maize, has played a crucial role in the genetic enhancement of maize. Several studies have highlighted the significant genetic diversity present in teosinte, which has been harnessed to improve various agronomic traits in maize. For instance, the introgression of the UPA2 allele from teosinte has been shown to enhance high-density maize yields by altering plant architecture to facilitate dense planting. Additionally, teosinte alleles have been identified that improve kernel composition traits such as starch, protein, and oil content, demonstrating the potential of teosinte to enhance the nutritional quality of maize. The teosinte branched1 (tb1) gene has been linked to the suppression of growth in maize, contributing to its less branched architecture compared to teosinte. Furthermore, the teosinte glume architecture1 (tga1) locus has been pivotal in the evolution of maize by reducing the hardness of glumes, making kernels more accessible for harvest. The discovery of a teosinte-derived allele of a MYB transcription repressor that confers multiple disease resistance in maize further underscores the value of teosinte in crop improvement.
The future of teosinte in maize genetic enhancement looks promising, with several avenues for further research and application. Advances in genomic and transcriptomic technologies, such as single-molecule long-read sequencing, have provided deeper insights into the genetic and transcriptomic variations between maize and teosinte, facilitating the identification of beneficial alleles for maize improvement. The development of robust genetic transformation protocols for teosinte, such as biolistic bombardment, opens up new possibilities for functional analyses of teosinte genes and their regulatory mechanisms. Additionally, the comprehensive characterization of the teosinte transcriptome has revealed adaptive sequence divergence during maize domestication, highlighting the potential of teosinte germplasm to enhance the adaptability of maize to various environmental stimuli. As we continue to explore the genetic potential of teosinte, it is likely that new alleles and genetic pathways will be discovered that can further enhance the yield, nutritional quality, and disease resistance of maize, ensuring food security in the face of growing global demands.
Acknowledgments
The CropSci Publisher extends sincere thanks to three anonymous peer reviewers for their feedback on the manuscript.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Adhikari S., Joshi A., Kumar A., and Singh N., 2021, Diversification of maize (Zea mays L.) through teosinte (Zea mays subsp. parviglumis Iltis and Doebley) allelic, Genetic Resources and Crop Evolution, 68: 2983-2995.
https://doi.org/10.1007/s10722-021-01170-z
Adhikari S., Joshi A., Kumar A., Singh N., Jaiswal J., Jeena A., and Pant U., 2022, Developing genetic resources and genetic analysis of plant architecture-related traits in teosinte-introgressed maize populations, Plant Genetic Resources: Characterization and Utilization, 20(2): 145-155.
https://doi.org/10.1017/s1479262122000223
Aguirre-Liguori J., Gaut B., Jaramillo‐Correa J., Tenaillon M., Montes-Hernández S., García-Oliva F., Hearne S., and Eguiarte L., 2019, Divergence with gene flow is driven by local adaptation to temperature and soil phosphorus concentration in teosinte subspecies (Zea mays parviglumis and Zea mays mexicana), Molecular Ecology, 28: 2814-2830.
https://doi.org/10.1111/mec.15098
PMid:30980686
Calfee E., Gates D., Lóránt A., Perkins M., Coop G., and Ross-Ibarra J., 2021, Selective sorting of ancestral introgression in maize and teosinte along an elevational cline, PLoS Genetics, 17(10): e1009810.
https://doi.org/10.1101/2021.03.05.434040
Chen Q., Samayoa L., Yang C., Bradbury P., Olukolu B., Neumeyer M., Romay M., Sun Q., Lóránt A., Buckler E., Ross-Ibarra J., Holland J., and Doebley J., 2020, The genetic architecture of the maize progenitor, teosinte, and how it was altered during maize domestication, PLoS Genetics, 16(5): e1008791.
https://doi.org/10.1371/journal.pgen.1008791
PMid:32407310 PMCid:PMC7266358
Ding S., Cai Z., Du H., and Wang H., 2019, Genome-wide analysis of tcp family genes in Zea mays L. identified a role for ZmTCP42 in drought tolerance, International Journal of Molecular Sciences, 20(11): 2762.
https://doi.org/10.3390/ijms20112762
PMid:31195663 PMCid:PMC6600213
Evans M., and Kermicle J., 2001, Teosinte crossing barrier1, a locus governing hybridization of teosinte with maize, Theoretical and Applied Genetics, 103: 259-265.
https://doi.org/10.1007/s001220100549
Fang H., Fu X., Wang Y., Xu J., Feng H., Li W., Xu J., Jittham O., Zhang X., Zhang L., Yang N., Xu G., Wang M., Li X., Li J., Yan J., and Yang X., 2019, Genetic basis of kernel nutritional traits during maize domestication and improvement, The Plant Journal, 101(2): 278-292.
https://doi.org/10.1111/tpj.14539
PMid:31529523
Fu Y., Xu G., Chen H., Wang X., Chen Q., Huang C., Li D., Xu D., Tian J., Wu W., Lu S., Li C., and Tian F., 2019, QTL mapping for leaf morphology traits in a large maize-teosinte population, Molecular Breeding, 39: 1-13.
https://doi.org/10.1007/s11032-019-1012-5
Gao H., Gadlage M., Lafitte H., Lenderts B., Yang M., Schroder M., Farrell J., Snopek K., Peterson D., Feigenbutz L., Jones S., Clair G., Rahe M., Sanyour-Doyel N., Peng C., Wang L., Young J., Beatty M., Dahlke B., Hazebroek J., Greene T., Cigan A., Chilcoat N., and Meeley R., 2020, Superior field performance of waxy corn engineered using CRISPR-Cas9, Nature Biotechnology, 38: 579-581.
https://doi.org/10.1038/s41587-020-0444-0
PMid:32152597
Gasca-Pineda J., Gutiérrez-Guerrero Y., Aguirre-Planter E., and Eguiarte L., 2020, The role of environment, local adaptation, and past climate fluctuation on the amount and distribution of genetic diversity in two subspecies of Mexican wild Zea mays, American Journal of Botany, 107(11): 1542-1554.
https://doi.org/10.1002/ajb2.1561
PMid:33205455
González J., Corral J., García G., Ojeda G., Larios L., Holland J., Medrano R., and Romero G., 2018, Ecogeography of teosinte, PLoS One, 13(2): e0192676.
https://doi.org/10.1371/journal.pone.0192676
PMid:29451888 PMCid:PMC5815594
Huang J., Gao Y., Jia H., and Zhang Z., 2016, Characterization of the teosinte transcriptome reveals adaptive sequence divergence during maize domestication, Molecular Ecology Resources, 16(6): 1465-1477.
https://doi.org/10.1111/1755-0998.12526
PMid:26990495
Hubbard L., McSteen P., Doebley J., and Hake S., 2002, Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte, Genetics, 162(4): 1927-1935.
https://doi.org/10.1093/genetics/162.4.1927
PMid:12524360 PMCid:PMC1462370
Joshi A., Adhikari S., and Singh N., 2021, Mapping genomic regions for red flour beetle (Tribolium castaneum (Herbst)) resistance in teosinte (Zea mays L. subsp. parviglumis H. H. Iltis and Doebley) derived maize backcross inbred line population, Genetic Resources and Crop Evolution, 68: 1529-1544.
https://doi.org/10.1007/s10722-020-01083-3
Karn A., Gillman J., and Flint-Garcia S., 2017, Genetic analysis of teosinte alleles for kernel composition traits in maize, G3: Genes|Genomes|Genetics, 7: 1157-1164.
https://doi.org/10.1534/g3.117.039529
PMid:28188181 PMCid:PMC5386864
Kumar A., Singh N., Adhikari S., and Joshi A., 2020, Morphological and molecular characterization of teosinte derived maize population, Indian Journal of Genetics and Plant Breeding, 79(4): 670-677.
https://doi.org/10.31742/IJGPB.79.4.4
Kumar A., Singh V., Saran B., Al‐Ansari N., Singh V., Adhikari S., Joshi A., Singh N., and Vishwakarma D., 2022, Development of novel hybrid models for prediction of drought-and stress-tolerance indices in teosinte introgressed maize lines using artificial intelligence techniques, Sustainability, 14(4): 2287.
https://doi.org/10.3390/su14042287
Li Z., Han L., Luo Z., and Li L., 2021, Single‐molecule long‐read sequencing reveals extensive genomic and transcriptomic variation between maize and its wild relative teosinte (Zea mays ssp. parviglumis), Molecular Ecology Resources, 22: 272-282.
https://doi.org/10.1111/1755-0998.13454
PMid:34157795
Liu L., Huang J., He L., Liu N., Du Y., Hou R., Du H., Qiu F., and Zhang Z., 2019, Dissecting the genetic architecture of important traits that enhance wild germplasm resource usage in modern maize breeding, Molecular Breeding, 39: 1-11.
https://doi.org/10.1007/s11032-019-1061-9
Mano Y., and Omori F., 2007, Breeding for flooding tolerant maize using "teosinte" as a germplasm resource, Plant Root, 1: 17-21.
https://doi.org/10.3117/PLANTROOT.1.17
Mano Y., and Omori F., 2013, Flooding tolerance in interspecific introgression lines containing chromosome segments from teosinte (Zea nicaraguensis) in maize (Zea mays subsp. mays), Annals of Botany, 112(6): 1125-1139.
https://doi.org/10.1093/aob/mct160
PMid:23877074 PMCid:PMC3783227
Mano Y., Omori F., Takamizo T., Kindiger B., Bird R., and Loáisiga C., 2006, Variation for root aerenchyma formation in flooded and non-flooded maize and teosinte seedlings, Plant and Soil, 281: 269-279.
https://doi.org/10.1007/s11104-005-4268-y
Rivera-Rodríguez D., Mastretta‐Yanes A., Wegier A., Larios L., Santacruz-Ruvalcaba F., Corral J., Hernández B., and González J., 2023, Genomic diversity and population structure of teosinte (Zea spp.) and its conservation implications, PLoS One, 18(10): e0291944.
Sahoo S., Adhikari S., Joshi A., and Singh N., 2021, Use of wild progenitor teosinte in maize (Zea mays subsp. mays) improvement: present status and future prospects, Tropical Plant Biology, 14: 156-179.
https://doi.org/10.1007/s12042-021-09288-1
Studer A., and Doebley J., 2012, Evidence for a natural allelic series at the maize domestication locus teosinte branched1, Genetics, 191: 951-958.
https://doi.org/10.1534/genetics.112.138479.
PMid:22505628 PMCid:PMC3389986
Tian J., Wang C., Xia J., Wu L., Xu G., Wu W., Li D. Qin W., Han X., Chen Q., Jin W., and Tian F., 2019, Teosinte ligule allele narrows plant architecture and enhances high-density maize yields, Science, 365: 658-664
https://doi.org/10.1126/science.aax5482
PMid:31416957
Wang H., Hou J., Ye P., Hu L., Huang J., Dai Z., Zhang B., Dai S., Que J., Min H., Chen G., Wang Y., Jiang M., Liang Y., Li L., Zhang X., and Lai Z., 2021, A Teosinte-derived allele of a MYB transcription repressor confers multiple disease resistance in maize, Molecular Plant, 14(11): 1846-1863.
https://doi.org/10.1016/j.molp.2021.07.008.
PMid:34271176
Wang K., Zhang Z., Sha X., Yu P., Li Y., Zhang D., Liu X., He G., Li Y., Wang T., Guo J., Chen J., and Li C., 2023, Identification of a new QTL underlying seminal root number in a maize-teosinte population, Frontiers in Plant Science, 14: 1132017.
https://doi.org/10.3389/fpls.2023.1132017
PMid:36824192 PMCid:PMC9941338
Warburton M., Wilkes G., Taba S., Charcosset A., Mir C., Dumas F., Madur D., Dreisigacker S., Bedoya C., Prasanna B., Xie C., Hearne S., and Franco J., 2011, Gene flow among different teosinte taxa and into the domesticated maize gene pool, Genetic Resources and Crop Evolution, 58: 1243-1261.
https://doi.org/10.1007/s10722-010-9658-1
Weber A., Briggs W., Rucker J., Baltazar B., Sánchez-Gonzalez J., Feng P., Buckler E., and Doebley J., 2008, The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): new evidence from association mapping, Genetics, 180: 1221-1232.
https://doi.org/10.1534/genetics.108.090134.
PMid:18791250 PMCid:PMC2567369
Xu G., Cao J., Wang X., Chen Q., Jin W., Li Z., and Tian F., 2019, Evolutionary metabolomics identifies substantial metabolic divergence between maize and its wild ancestor, teosinte, Plant Cell, 31: 1990-2009.
https://doi.org/10.1105/tpc.19.00111
PMid:31227559 PMCid:PMC6751114
Yang C., Samayoa L., Bradbury P., Olukolu B., Xue W., York A., Tuholski M., Wang W., Daskalska L., Neumeyer M., Sanchez-Gonzalez J., Romay M., Glaubitz J., Sun Q., Buckler E., Holland J., and Doebley J., 2019, The genetic architecture of teosinte catalyzed and constrained maize domestication, Proceedings of the National Academy of Sciences of the United States of America, 116: 5643-5652.
https://doi.org/10.1073/pnas.1820997116
PMid:30842282 PMCid:PMC6431195
Zavala-López M., López-Tavera E., Figueroa-Cárdenas J., Serna-Saldívar S., and García‐Lara S., 2018, Screening of major phenolics and antioxidant activities in teosinte populations and modern maize types, Journal of Cereal Science, 79: 276-285.
https://doi.org/10.1016/J.JCS.2017.11.007
Zhang X., Lu M., Xia A., Xu T., Cui Z., Zhang R., Liu W., and He Y., 2021, Genetic analysis of three maize husk traits by QTL mapping in a maize-teosinte population, BMC Genomics, 22: 1-9.
https://doi.org/10.1186/s12864-021-07723-x
PMid:34034669 PMCid:PMC8152318
Zobrist J., Martin-Ortigosa S., Lee K., Azanu M., Ji Q., and Wang K., 2021, Transformation of tosinte (Zea mays ssp. parviglumis) via biolistic bombardment of seedling-derived callus tissues, Frontiers in Plant Science, 12: 773419.
https://doi.org/10.3389/fpls.2021.773419
PMid:34956270 PMCid:PMC8696365

. PDF(639KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Shanjun Zhu
. Wei Wang
Related articles
. Teosinte
. Genetic diversity
. Maize improvement
. Alleles
. Sustainable agriculture
Tools
. Email to a friend
. Post a comment